Recursive Multivariate Testing for Cohort Clustering with Disparate Covariate Information
Source:vignettes/relate.Rmd
relate.RmdData setup
set.seed(123)
#load simulated dataset from the package
data("cohort_na_df")
cohort_na_df[199:202,]
#> X1 X2 X3 X4 X5 X6 X7
#> 199 9.800547 11.192049 13.22608 11.741157 3.2985284 17.568287 16.80876
#> 200 9.931415 10.138727 11.55728 13.071213 0.1115508 9.439059 12.11496
#> 201 4.040424 10.757824 NA 5.497721 -2.1834396 -7.249764 16.84709
#> 202 4.658563 8.917538 NA 5.997931 1.5099440 8.548666 11.56883
#> X8 X9 X10 X11 X12 X13 X14
#> 199 19.57499 15.711291 8.704099 14.994329 19.08767 17.18427 9.340641
#> 200 19.50130 15.995014 12.911449 8.116613 20.50924 17.98611 2.671327
#> 201 11.47614 10.095340 NA 7.683143 24.87351 12.84377 7.324220
#> 202 16.18313 8.848892 NA 10.561502 18.80442 19.89488 8.512655
#> X15 cohortid distribution
#> 199 12.6633576 2 1
#> 200 9.7992329 2 1
#> 201 1.0246224 3 1
#> 202 0.8836475 3 1
#save the true distribution
true_distribution <- cohort_na_df$distribution
cohort_na_df <- cohort_na_df %>% dplyr::select(-distribution)
# examine missing pattern (e.g., some cohorts did not collect specific information)
cohort_na_df %>% find_na_level(clusterid = "cohortid")
#> # A tibble: 10 × 16
#> ClusterID X1 X2 X3 X4 X5 X6 X7 X8 X9 X10 X11
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0 0 0 0 0 0 0 0 0 0 0
#> 2 2 0 0 0 0 0 0 0 0 0 0 0
#> 3 3 0 0 1 0 0 0 0 0 0 1 0
#> 4 4 0 0 0 0 0 0 0 0 0 0 0
#> 5 5 0 0 0 1 0 0 0 0 0 0 0
#> 6 6 0 0 0 0 0 0 0 0 0 0 0
#> 7 7 0 0 0 0 0 0 0 0 1 0 1
#> 8 8 0 0 0 0 0 0 0 0 0 0 0
#> 9 9 0 0 1 0 1 0 0 0 0 0 0
#> 10 10 0 0 0 0 0 0 0 0 0 0 0
#> # … with 4 more variables: X12 <dbl>, X13 <dbl>, X14 <dbl>, X15 <dbl>
#> # ℹ Use `colnames()` to see all variable names
#add random missingness in addition to the systematic missingness for a more realistic scenario
random_missing_row <- sample(1:nrow(cohort_na_df), floor(nrow(cohort_na_df)/2))
missing_col <- c(1,8,13,14)
cohort_na_df[random_missing_row, missing_col] <- NA
# examine missing pattern after adding random missingness
cohort_na_df %>% find_na_level(clusterid = "cohortid")
#> # A tibble: 10 × 16
#> ClusterID X1 X2 X3 X4 X5 X6 X7 X8 X9 X10 X11
#> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 0.49 0 0 0 0 0 0 0.49 0 0 0
#> 2 2 0.55 0 0 0 0 0 0 0.55 0 0 0
#> 3 3 0.48 0 1 0 0 0 0 0.48 0 1 0
#> 4 4 0.54 0 0 0 0 0 0 0.54 0 0 0
#> 5 5 0.49 0 0 1 0 0 0 0.49 0 0 0
#> 6 6 0.47 0 0 0 0 0 0 0.47 0 0 0
#> 7 7 0.49 0 0 0 0 0 0 0.49 1 0 1
#> 8 8 0.48 0 0 0 0 0 0 0.48 0 0 0
#> 9 9 0.53 0 1 0 1 0 0 0.53 0 0 0
#> 10 10 0.48 0 0 0 0 0 0 0.48 0 0 0
#> # … with 4 more variables: X12 <dbl>, X13 <dbl>, X14 <dbl>, X15 <dbl>
#> # ℹ Use `colnames()` to see all variable namesConduct unsupervised random forest using cohort id as the outcome
analytic.rfsrc <- rfsrc.fast(cohortid ~ ., #regress cohort id on all other variables
data = cohort_na_df,
distance=T, ntree=1500, nsplit=3,
na.action="na.impute",nimpute=1) # impute within nodes
distance.matrix <- analytic.rfsrc$distance # extract distance matrix Hierachical clustering of the average distance matrix
#calculate average distance matrix
dist_avg_mat <- avg_dist_func(distance.matrix, cohort_na_df$cohortid)
#cluster average distance matrix and
#identify which cohorts are clustering together on dendogram
dmean <- as.dist(dist_avg_mat)
#hierarchical clustering
hc2 <- hclust(dmean, method = "ward.D2")
dend <- as.dendrogram(hc2)
plot(dend)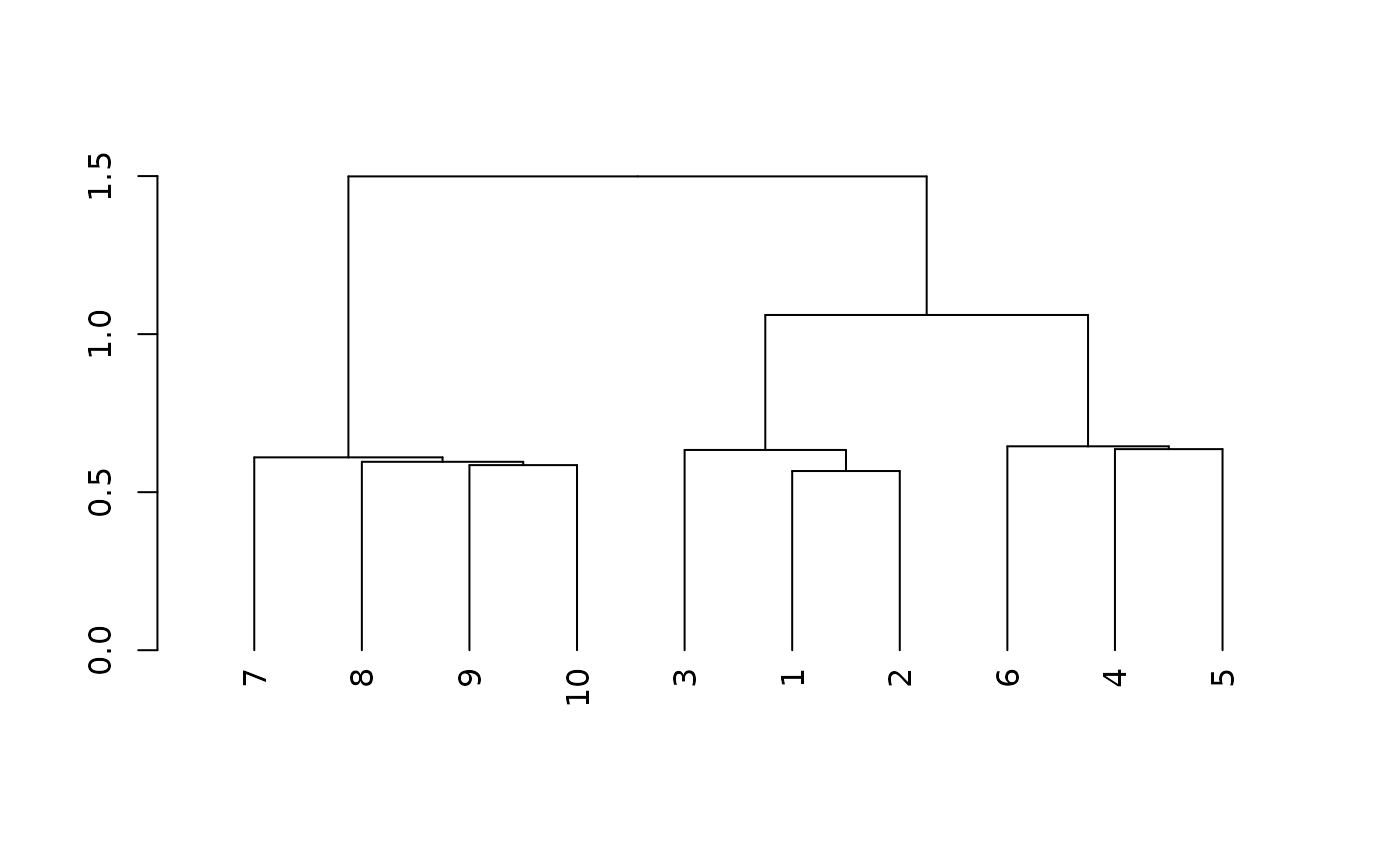
Within cohort imputation to account for Missing At Random (MAR) variables
We generally do not impute the missing variables that are completely missing or missing more than 50%.
The relate::mice_group_impute in this package provides imputation by group regardless of missingness level (imputeLevel = 1). Please find miceadds::mice.impute.bygroup for similar functionality.
mice_group_impute(df = cohort_na_df, clusterid = "cohortid", imputeLevel = 1, miceArg = list(method = 'mean', maxit = 1))Recursive BG testing based on the previous dendrogram
# we use mean imputation and asymptotic BG test for illustrative purposes.
ll <- recursive.test(dend = dend, df = cohort_na_df, cohortid.var = "cohortid",
verbose = T, saveIntermediate = F,
BG.method = 'permutation', n_perm = 100, impute = T,
miceArgs = list(method = c("mean"), m =1, maxit = 5, seed = 123, printFlag = F))
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 9 10
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.49 FALSE
#> --------------------------
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 8 9,10
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.29 FALSE
#> --------------------------
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 7 8,9,10
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.03 TRUE
#> --------------------------
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 1 2
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.32 FALSE
#> --------------------------
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 3 1,2
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.71 FALSE
#> --------------------------
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 4 5
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.79 FALSE
#> --------------------------
#> Start testing:
#> # A tibble: 1 × 2
#> X Y
#> <chr> <chr>
#> 1 6 4,5
#> Results:
#> # A tibble: 1 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0.04 TRUE
#> --------------------------
#> Start testing:
#> # A tibble: 2 × 2
#> X Y
#> <chr> <chr>
#> 1 3,1,2 6
#> 2 3,1,2 4,5
#> Results:
#> # A tibble: 2 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0 TRUE
#> 2 0 TRUE
#> --------------------------
#> Start testing:
#> # A tibble: 6 × 2
#> X Y
#> <chr> <chr>
#> 1 7 3,1,2
#> 2 8,9,10 3,1,2
#> 3 7 6
#> 4 8,9,10 6
#> 5 7 4,5
#> 6 8,9,10 4,5
#> Results:
#> # A tibble: 6 × 2
#> Pairwise.p Significant
#> <dbl> <lgl>
#> 1 0 TRUE
#> 2 0 TRUE
#> 3 0 TRUE
#> 4 0 TRUE
#> 5 0 TRUE
#> 6 0 TRUE
#> --------------------------
ll$clusters
#> [1] "7" "8,9,10" "3,1,2" "6" "4,5"
table(true_distribution, cohort_na_df$cohortid, dnn = c("True Cluster","Cohort ID"))
#> Cohort ID
#> True Cluster 1 2 3 4 5 6 7 8 9 10
#> 1 100 100 100 0 0 0 0 0 0 0
#> 2 0 0 0 100 100 100 0 0 0 0
#> 3 0 0 0 0 0 0 100 100 100 100